Bimodal Gene Expression in Breast Cancer
-
Jessica C. Mar, PhD
Assistant Professor, Department of Systems & Computational Biology, Assistant Professor, Department of Epidemiology & Population Health, Albert Einstein College of MedicineBIOGRAPHY
Tumors are often categorized into standard molecular subtypes. However, largescale studies have demonstrated that patient heterogeneity in the regulatory make-up of tumors remain. At the transcriptional level, one example of heterogeneity in a patient population is the presence of bimodally-expressed genes. Bimodality in expression signifies the presence of potentially new patient sub-groups. Here, we present a new statistical approach called oncomix, that models transcriptional heterogeneity in tumor and adjacent normal (i.e. tumor-free) using bimodality to find oncogene candidates. Oncomix was applied to RNA-sequencing data from the breast cancer cohort of the Cancer Genome Atlas, and a set of oncogene candidates that were over-expressed in only a subset of tumors was identified.
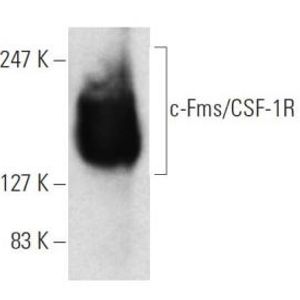
Intronic DNA methylation was strongly associated with the overexpression of chromobox 2 (CBX2), an oncogene candidate that was identified using our method but not through other approaches. CBX2 overexpression in breast tumors was associated with the upregulation of genes involved in cell cycle progression and is associated with poorer 5-year survival. The predicted function of CBX2 was confirmed in vitro providing the first experimental evidence that CBX2 promotes breast cancer cell growth. Modeling mRNA expression heterogeneity in tumors through bimodal profiles is a novel powerful approach with the potential to uncover therapeutic targets that benefit subsets of cancer patients.
Bimodal Gene Expression in Breast Cancer
Please update your information
Certificate of Attendance
DOWNLOAD CERTIFICATE
Finish Registering
-
APR 17, 2024Cannabis Sciences Virtual Event Series 2024
-
APR 30, 2024Immuno-Oncology Virtual Event Series 2024
-
MAY 07, 20243rd International Biosecurity Virtual Symposium
- See More
-
APR 15, 2024
-
APR 15, 2024
- See More