Learn How Spatial Biology Can Advance Your Research
To meet the demands of cutting-edge biomedical research, NanoString have launched a spatial transcriptomics platform called the GeoMx® Digital Spatial Profiler (DSP), which is validated with Illumina's next-generation sequencing platforms. This integrated solution aims to help researchers learn more about when, where and at what levels genes are expressed in cells or tissues of interest.
Understanding physiological processes and treating disease often requires detailed knowledge about gene expression. Over the years, a variety of tools and techniques have been developed so that scientists can investigate how, why, when, and where gene and protein expression occur in different cell and tissue types. But researchers are beginning to reach the limits of some of those methods, many of which are focused on only a small number of proteins or genes at once instead of the many molecules that are active in a cell at the same time.
For example, cells can be treated in many ways with fluorescent antibodies or other reagents that colorfully highlight specific RNAs or proteins of interest in a tissue, but because of the color spectrum, there are only so many of them that can be highlighted at the same time.
There are other drawbacks to staining cells as well. To see what's happening inside of cells or tissue samples, many of these techniques require fixation of cells or tissue. While staining techniques like immunofluorescence or in situ hybridization that usually require fixation can tell a researcher more about when or where a protein is expressed, they are unable to provide quantitative data about the level of molecules, or information about a live organism.
Live cells can be imaged to see molecules, but they often rely on genetic engineering tools that tag proteins with a fluorescent protein at the genomic level, so they will glow when the cell expresses them. Yet, again, the number of proteins that can be tagged simultaneously is limited. It also takes extra time to incorporate these genetic edits in the targeted cells or organisms. Despite being small sizes, the tags introduced may interfere with the natural function of genes and proteins.
A popular technology in the recent years is Single-cell sequencing, which is a next-generation sequencing (NGS) method that examines the genomes or transcriptomes of individual cells by providing a high-resolution view of cell-to-cell variation. This technology enables researchers to explore the distinct biology of individual cells in complex tissues and understand cellular subpopulation responses to environmental cues. While Single-cell sequencing analysis allows scientists to measure all the gene activities at cellular resolution, it is unable to map the locations of activities occurring within the tissue.
Spatial biology is an emerging field that aims to overcome the challenges discussed above and improves our understanding of gene activity in a tissue sample with morphological context. The goal of spatial biology is to take the investigator to the molecular level of a sample and obtain data that can't be gleaned from other techniques. Data can be collected about the genes that are expressed in a cell at one time, but spatial biology methods also show where the messenger RNA transcripts of active genes are located. Genes can be seen within the context of other genes, instead of looking at them in isolation or with only a few other cell markers. This is of particularly interest to researchers who like to investigate the spatial distribution of cells and their neighboring microenvironment.
"Spatial transcriptomics will allow us to understand gene expression in the context of the spatial location of each cell. So, we can see the different interactions between immune cells and tumor cells, and also what signaling pathways are going on at different regions of the tumor," said Dr. Vivian Lim, a Research Fellow at the Institute of Molecular and Cell Biology in Singapore.
In 2020, Nature Methods declared that spatial transcriptomics was the “Method of the Year.” When Nature Methods described the technique, they quoted researchers who expect it to be an important research tool in coming years, just like single-cell gene expression analysis has opened up a new understanding of physiology. Spatial transcriptomic tools are already being used to build cell atlases. It may one day be possible to use these techniques in live tissues as well. With a vast range of potential applications, this technique is currently being applied to many different fields including neurobiology, oncology, and COVID-19 studies, enabling researchers to obtain an unprecedented amount of information from a single tissue sample.
One of the leading spatial transcriptom platform is developed by NanoString. GeoMx DSP was first launched in 2019 and it has been brought together with Illumina's next-generation sequencing platforms to create the end-to-end spatial transcriptomics solution, with the motto as “any target, any region, any sample.” This solution empowers the researchers to analyze biomarkers in samples so they can visualize the activity of genes and proteins in the spatial context.
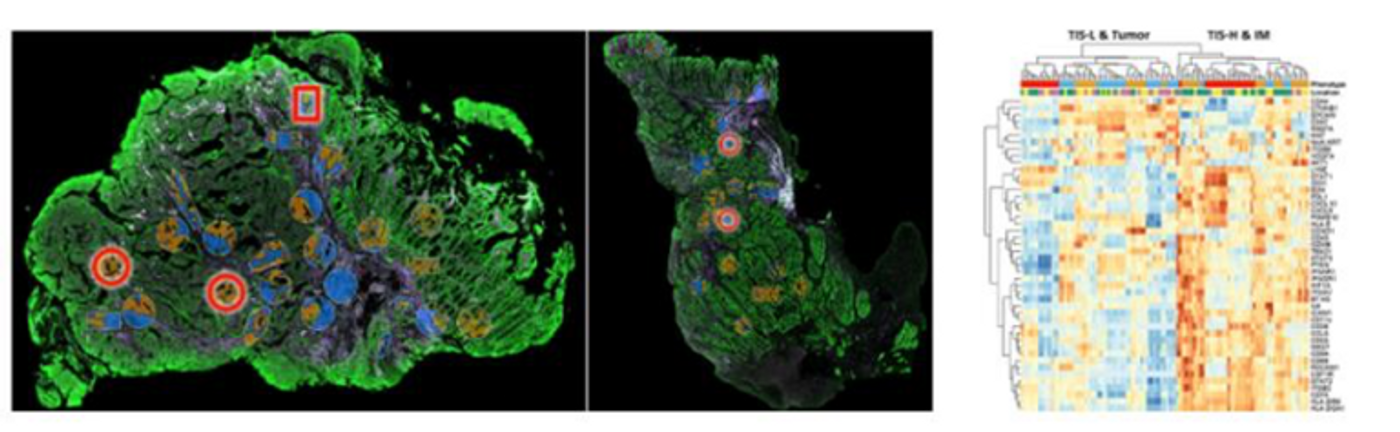
In this method, a mixture oligonucleotide probes first bind to their mRNA targets. The plex number of these oligonucleotide probes can range from a custom set under 100 to more than 18,000 in human. Each of the probes are connected to a unique barcode sequence along with a linker that can be deactivated upon exposure to UV light. With the accompanying imaging software in GeoMx DSP, researchers can specify the region of interest (ROI) with guidance of fluorescent proteins staining. By illuminating the region of interest (ROI) in the tissue with UV light, the barcodes are released and collected for further analysis. The ROI selection is highly flexible and can be performed even on Tissue Microarray (TMA)
One of the major benefits is that GeoMx DSP technology has been proven to work excellently with tissues that have been chemically fixed with formalin, and then embedded in paraffin. Formalin-fixed, paraffin-embedded or FFPE, is a common way to preserve tissues in clinic settings and biobanks. It is also the major resource of research samples in translational and clinical science. In most of the clinical setting, the tissue block can be sliced into thin sections and stained for proteins of interest. Due to the nature of preservation, it has been challenging for researchers to profile gene expression in these tissues. With NanoString's GeoMx DSP and Illumina’s NGS now, these highly processed tissues can be evaluated with spatial transcriptomics at whole transcriptome level to get a more complete picture of the biology at work.
The captured oliogonucleotides are converted to NGS libraries with adapters compatible with Illumina sequencers. Researchers can set up runs on Illumina instruments with data automatically uploaded onto Illumina’s BaseSpaceTM Sequence Hub (BSSH). Once the data is in available in BSSH (in a few mins after the sequencing is complete), the researcher can select the DRAGENTM (Dynamic Read Analysis for GENomics) accelerated GeoMx NGS Pipeline from the Apps menu for data analysis. Analysis will typically complete in 1 to 2 hours, for a typical experiment containing ~192 regions of interest (ROIs) and ~400 million reads. After the analysis completes, customers can download the results, and load them into their GeoMx DSP software for interactive visualization.
"It will take about 12-13 hours for the sequencing to be done, and then all the data will be uploaded onto BaseSpace," said Dr. Lim. "From sample prep all the way through to the data analysis, everything is very user-friendly and streamlined."
But it’s very simple to use. "Just load the sample - put it in the machine and click 'Start,'" said Dr. Timothy Shuen, a Research Fellow National Cancer Center Singapore about his experience of sequencing the spatial transcriptomics libraries on Illumina NextSeqTM 2000 platform.
"We can use our BaseSpace account to analyze and assess the data. So, to me it's really straightforward and seamless,” said Dr. Shuen. “The integrated solution of NanoString DSP technology, Illumina sequencing, and the BaseSpace analytics solution is very impressive."
As of April 2021, Illumina made this application available to users globally who have an active BaseSpace Sequence Hub Professional subscription. New customers can try BaseSpace for free for thirty days to see how this powerful technology can accelerate their research.
-Written by Labroots Science Writer Carmen Leitch
Research Use Only. Not for use in diagnostic procedures.
Sources:
https://www.nanostring.com/products/geomx-digital-spatial-profiler/geomx-dsp-overview/