CRISPR technology continues to revolutionize biology; in two new papers published in Science, researchers have revealed new techniques that turn the gene editor into a diagnostic tool. The first, called DETECTR, can identify bits of DNA that indicate the presence of viral infections or cancer growth. It may also be useful for finding defective genes. The other tool, termed SHERLOCK, is a paper-based diagnostic test that can identify genetic signatures or target molecules associated with myriad health conditions, like pregnancy for just one example. The researchers believe these tools will have a vast range of applications.
"SHERLOCK provides an inexpensive, easy-to-use, and sensitive diagnostic method for detecting nucleic acid material - and that can mean a virus, tumor DNA, and many other targets," said senior author Feng Zhang, who was central to the development of the original CRISPR gene-editing technology. He is a core institute member at the Broad Institute, an investigator at the McGovern Institute for Brain Research at MIT, and the James and Patricia Poitras Professor in Neuroscience at MIT. "The SHERLOCK improvements now give us even more diagnostic information and put us closer to a tool that can be deployed in real-world applications."
The investigators have already demonstrated the power of SHERLOCK, and in their new paper, they show that it can be used to find cell-free tumor DNA present in blood taken from lung cancer patients. It can also identify Zika and Dengue viruses simultaneously. The team wants to develop something that can easily be used in the field.
"The new paper readout for SHERLOCK lets you see whether your target was present in the sample, without instrumentation," said co-first author Jonathan Gootenberg, a graduate candidate in Zhang's lab and the lab of Broad core institute member Aviv Regev. "This moves us much closer to a field-ready diagnostic."
SHERLOCK uses Cas13, which scientists can program, so it binds to specific pieces of target RNA, whether it's a gene that makes a bacterium resistant to antibiotics, a snippet of viral DNA, or a cancer-causing mutation. Once it finds the genetic sequence it is zeroing in on, it chops up pieces of synthetic RNA, added to act as a signal to show that Cas13 has found its target.
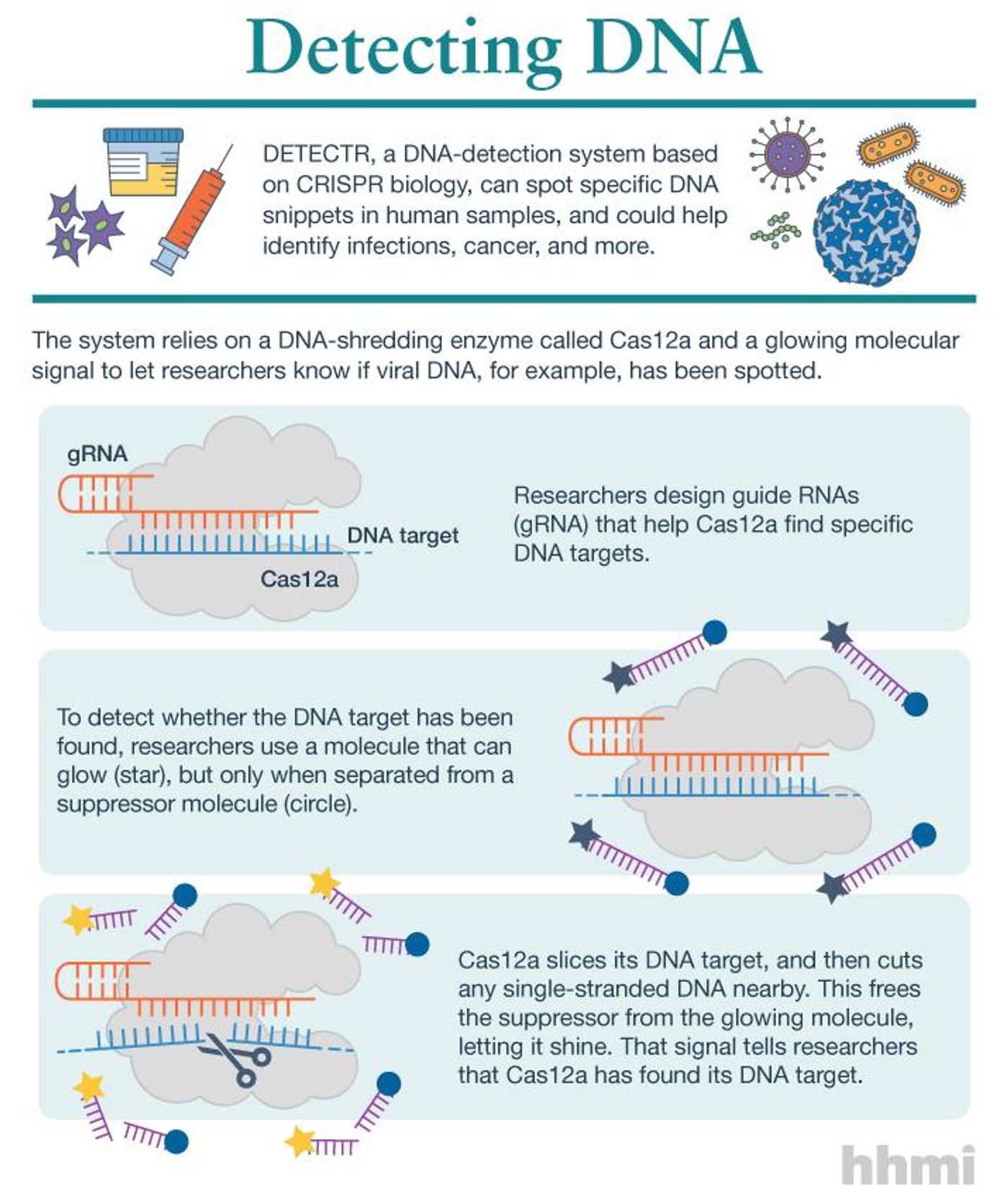
In the DETECTR system, an enzyme called Cas12a was utilized. It can be reprogrammed to find specific pieces of DNA as well. It also cuts DNA like other Cas enzymes, but sometimes it chops up not only the DNA it has bound to but other DNA as well. "We started to see this surprising activity where it would just start cutting random stuff," explained one author of the new report, Lucas Harrington, a graduate student in the lab of Jennifer Doudna. She was another inventor of the original CRISPR gene-editing tool.
They tweaked the enzyme so it would recognize a target, but then would turn to a piece of added DNA which has a glowing molecule linked to a suppressor strand. When Cas12a finds its target and binds to it, it then slices the DNA that connects the signal to the suppressor, and the glowing molecule lights up as an indicator.
The team tested their creation on the diagnosis of two types of HPV. When it came to HPV16, DETECTR made the right diagnosis for every one of 25 samples that were tested. For HPV18, the DETECTR system was correct 23 out of 25 times. That makes it a much easier, faster method that doesn't require special equipment. The researchers think they can improve HPV18 detection to be totally accurate.
Now this work can be used for other diseases and diagnostics. CRISPR "is a treasure chest that we keep digging into and finding new things," Harrington concluded.
Zhang is featured in the video above, discussing how CRISPR can help detect disease.
Sources: AAAS/Eurekalert!, HHMI, Broad Institute, Science Gootenberg et al, Science Chen at al