Investigating the Birth of Mitochondria
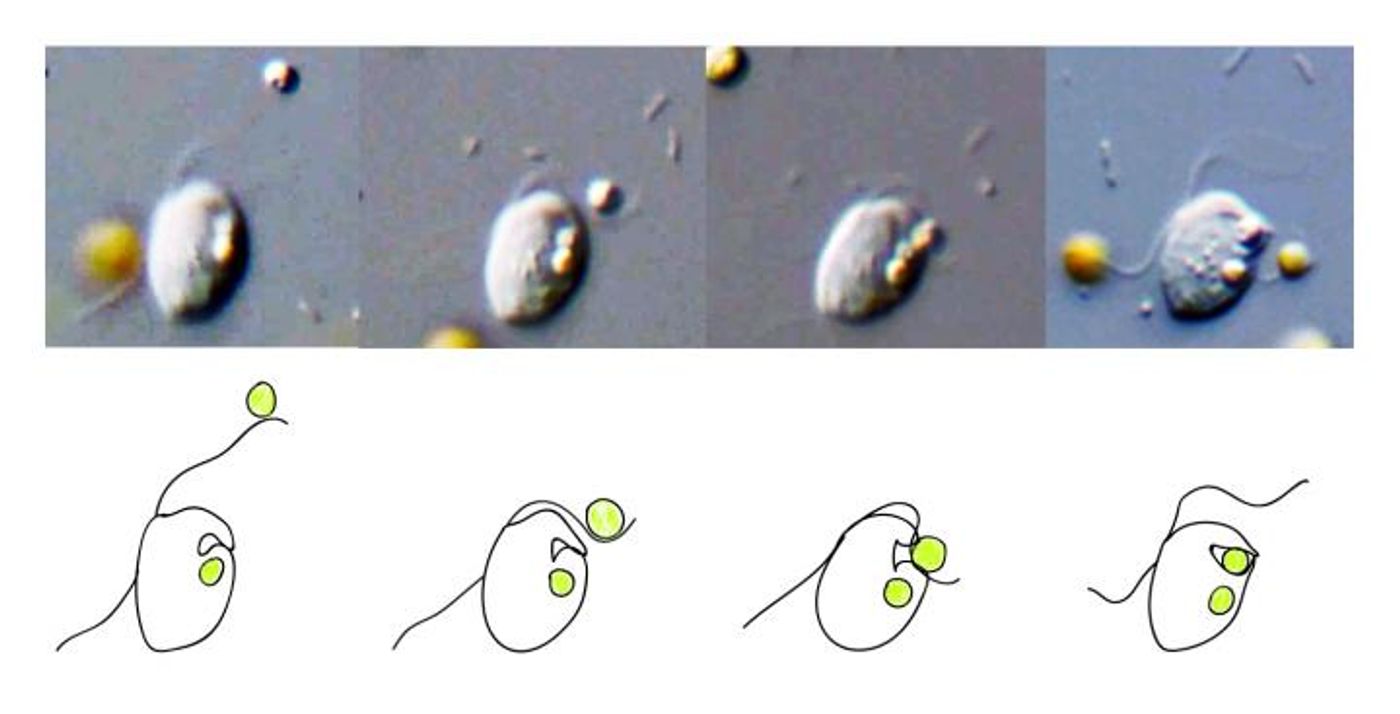
In a process called phagocytosis, a cell ingests another cell, and that is thought to have played a role in evolution. Now researchers at the American Museum of Natural History have created a computer program that can predict whether a cell can phagocytose another. The tool will be useful to scientists studying how complex organisms arose, and provides insight into microbial assessments. The work has been reported in Nature Ecology & Evolution.
"Phagocytosis is a major mechanism of nutritional uptake in many single-celled organisms, and it's vital to immune defenses in a number of living things, including humans," noted study co-author Eunsoo Kim, an associate curator in the American Museum of Natural History's Division of Invertebrate Zoology. "But lesser known is the idea that phagocytosis dates back some two to three billion years and played a role in those symbiotic associations that likely started the cascading evolution toward the more diverse and complex life we see on the planet today. Our research provides some hints as to how phagocytosis first arose."
Prokaryotes are microscopic, primarily single-celled organisms, and include bacteria and archaea. Animals, protists, plants, and fungi are eukaryotes, with cells that are typically larger than prokaryotes and are divided into organelles. Those include the mitochondria, a powerhouse, and the nucleus, which contains the genetic material. It has been theorized that about two or three billion years ago, a single-celled bacterium merged with an unrelated microbe, leading to the organelle we now know as the mitochondria and a critical feature of eukaryotes. That event only happened one time though, and there aren’t any known prokaryotes can engulf other cells.
The investigators wanted to investigate the question of how that merger occurred. They utilized patterns in the genetics of phagocytic cells to engineer a model that can use machine learning to identify organisms that ingest food by phagocytosis.
"There's no single set of genes that are strongly predictive of phagocytosis because it's a very complicated process that can involve more than 1,000 genes, and those genes can greatly vary from species to species," explained lead author John Burns, a research scientist in the Museum's Sackler Institute for Comparative Genomics." But as we started looking at the genomes of more and more eukaryotes, a genetic pattern emerged, and it exists across diversity, even though it's slightly different in each species. That pattern is the core of our model, and we can use it to very quickly and efficiently predict which cells are likely to be 'eaters' and which are not."
Some eukaryotes are picky eaters, and can’t be easily observed eating; instead, researchers can use this model to help identify organisms that can phagocytose. Check out the phagocytosis of bacteria by macrophages, immune cells, in the video.
In this study, the scientists used their model to study a group of microbes that are similar to eukaryotes, called Asgard archaea. It has been suggested that these microorganisms may be related to the ones that merged with the bacterium that evolved into mitochondria. This study found, however, the Asgard archaea probably don’t phagocytose.
In addition, the researchers checked the gene list used to create their model. Comparing it with ancient strains of bacteria and archaea, they think neither type of organism carries all of the genes needed for phagocytosis. While Asgard archaea are out of the candidate list, many remain.
"When you tease apart the components of these predictive genes, some have roots in archaea, some have roots in bacteria, and some are only unique to eukaryotes," Kim explained. "Our data are consistent with the hypothesis that our cells are a chimera of archaeal and bacterial components, and that the process of phagocytosis arose only after that combination took place. We still have a lot of work to do in this field."
Sources: AAAS/Eurekalert! Via American Museum of Natural History, Nature Ecology & Evolution