Cell Line Authentication Using STR Analysis
Imagine you’re studying colon cancer using a colon cell line model. After three painstaking years of research, your first major publication describes several new findings on the molecular pathways in this model. Or perhaps you identify sensitivity to a particular drug treatment. All of this work—your findings, its effect on the medical community, your reputation and grant access—hinge on your understanding of this cell line as a colon cancer model. But what happens if that cell line, gifted from the lab across the hall, has been mislabeled during routine passage? Or was overtaken by the faster growing HeLa cells also cultured in your lab?
Cell mislabeling or contamination is such a rare occurrence, you don’t have to worry about that, right? Unfortunately, misidentified cells are all too common. In 1999, an estimated 18% of cell lines submitted to a German cell bank were misidentified (1). The Web of Science database uncovered 32,755 research articles that use cell lines known to be misidentified (2). And the TRIP Lab, a core facility providing cell line authentication, estimated that 28% of cell lines tested in 2017 were either contaminated or misidentified (3). These numbers are startling, and certainly not something you want to bet your career or the lives of patients on.
But there is good news, too. Major granting institutions and many publications are now requiring cell line authentication. Awareness of this issue is improving in the research community. And this shift in awareness and funding is having an effect. After outreach to researchers in its institution, the TRIP Lab showed that misidentified and contaminated cell lines dropped from 28% to 3.8% in just 2 years (3).
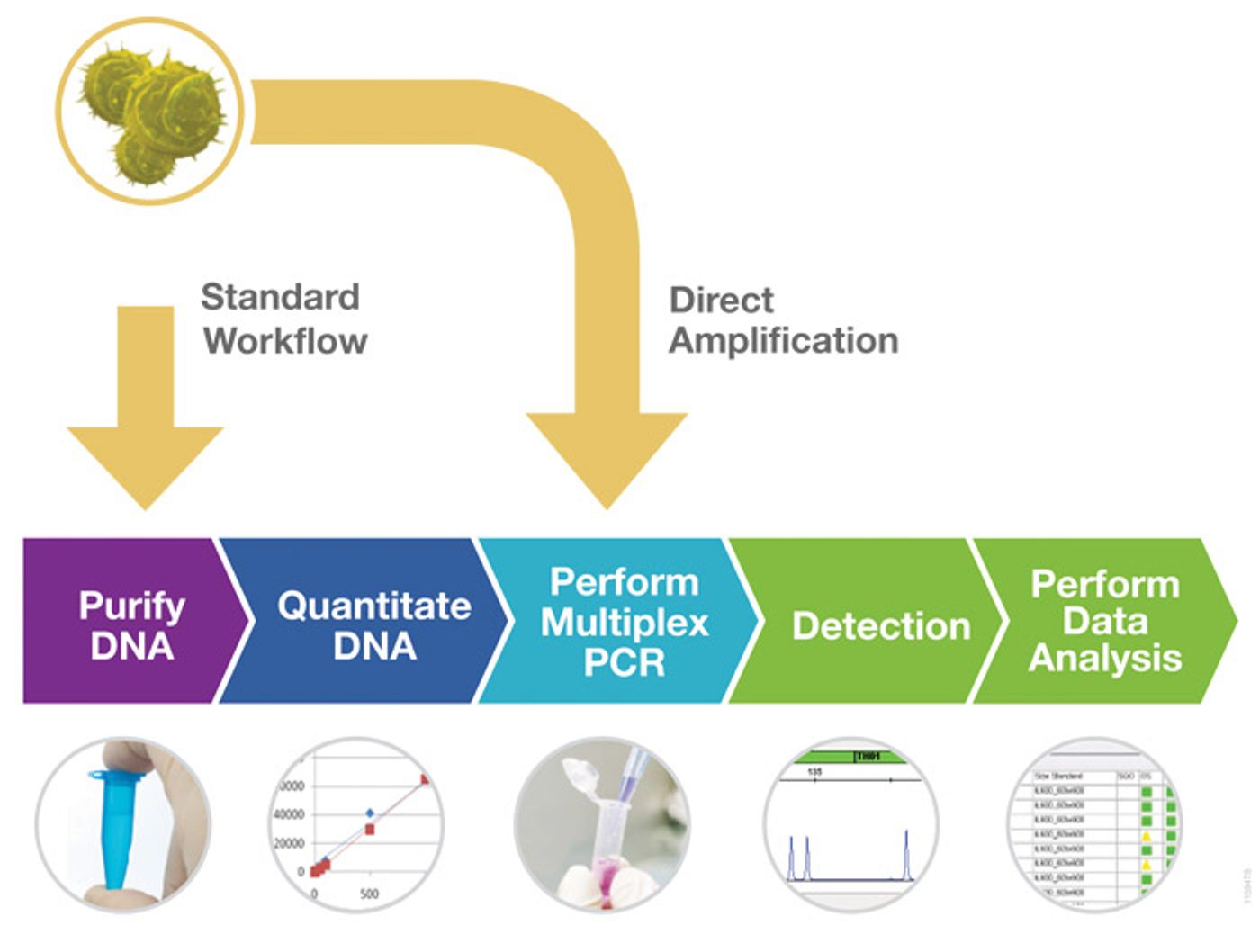
To deal with the problem of misidentified or contaminated cells, a committee of experts published the ANSI/ATCC ASN-0002-2011 consensus guidelines for best practices in cell line authentication based on STR genotyping (4). Short tandem repeats (STRs) are 2–7bp repeating DNA sequences in the genome that are typically highly polymorphic between individuals in a population. These regions can be amplified by PCR using primers outside of the repeat sequence, and the resulting amplicons sized using capillary electrophoresis to determine the number of repeats for each individual. This technology is commonly used in the forensic community for identification of individuals, and can also be used to distinguish cell lines from different human donors. Most STR chemistries currently available can be performed as multiplexes and use multiple dye channels for electrophoresis for concurrent analysis of many STR loci at the same time.
Following the publication of the ANSI/ATCC guidelines, the International Cell Line Authentication Committee (ICLAC) was established to maintain a register of cross-contaminated and misidentified cell lines. Their website is also an excellent collection of resources for cell line authentication (CLA), including links to several current STR databases, guides and advice for individual scientists, and policy recommendations for institutions seeking to establish rigorous science with authenticated cell lines as the norm (5).
Since cell line authentication is expected for grant applications submitted to the NIH and required for publication in many scientific journals, it is important to become familiar with the basic principles and best practices. To learn more about these steps and how to perform them using STR analysis on the Spectrum Compact CE System, download the application note: Cell Line Authentication Using the GenePrint® 10 and GenePrint® 24 Systems