New Method IDs Novel Pathogen Variants in Real Time
New technologies like wastewater surveillance have enabled public health officials to monitor the spread of a few infectious diseases in a community by identifying known pathogens by their DNA. While this tool has been helpful, it does not usually reveal variants of bacterial or viral pathogens (except in the case of flu or COVID), or emerging instances of antibiotic resistance. Scientists have now crated a new tool that can do this with patient samples. The researchers who developed it called it Phylowave.
The work has been reported in Nature, and it could enable the real-time monitoring of infections in large groups or populations of people. It may also help scientists who are creating medicines or vaccines, or aid epidemiologists and other researchers who are investigating why some variants spread while others do not.
This tool relies on genetic sequencing data that can show when new variants are emerging. One potential limitation of the technology is that it relies on samples that are obtained from people with active infections. But that also means that it can be applied in places with few resources, which is a great thing.
This technique can rapidly reveal if new, transmissible pathogen variants are circulating in a population, for a range of bacteria and viruses, noted corresponding study author Dr. Noémie Lefrancq, now of ETH Zurich. “We can even use it to start predicting how new variants are going to take over, which means decisions can quickly be made about how to respond.”
This "completely objective" tool "can rapidly and effectively spot the emergence of new highly transmissible strains,” added study co-author Professor Julian Parkhill, a researcher at the University of Cambridge.
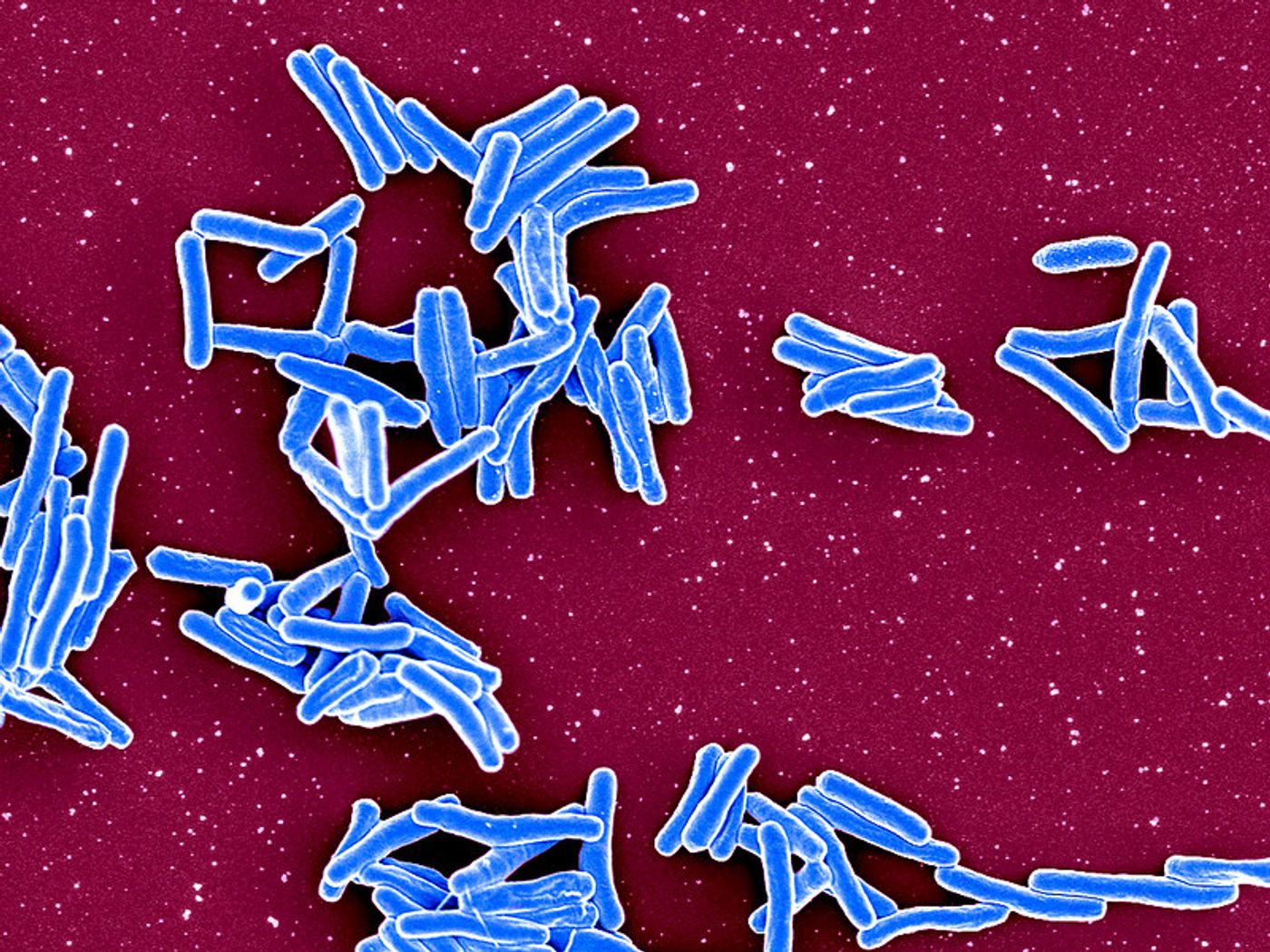
In this work, the investigators tested Phylowave with the pathogen that causes whooping cough: Bordetella pertussis. There are serious whooping cough outbreaks in several countries around the world right now. Phylowave found three new variants of this pathogen that had not been detected before this.
The video above describes how advanced genetic sequencing tools can help identify emerging pathogens and novel variants.
Phylowave was also used to assess samples of the pathogen that causes tuberculosis, known as Mycobacterium tuberculosis. Two antibiotic-resistant variants of M. tuberculosis were identified.
In some cases, these variants may be targetable with a newly designed vaccine. “If we see a rapid expansion of an antibiotic-resistant variant, then we could change the antibiotic that’s being prescribed to people infected by it, to try and limit the spread of that variant,” said senior study author Professor Henrik Salje of the University of Cambridge.
The study authors envision this technology as one facet of a larger effort that can better protect public health as pathogens continue to emerge, circulate, and change.
Sources: University of Cambridge, Nature