A Better Way to Classify Bacteria
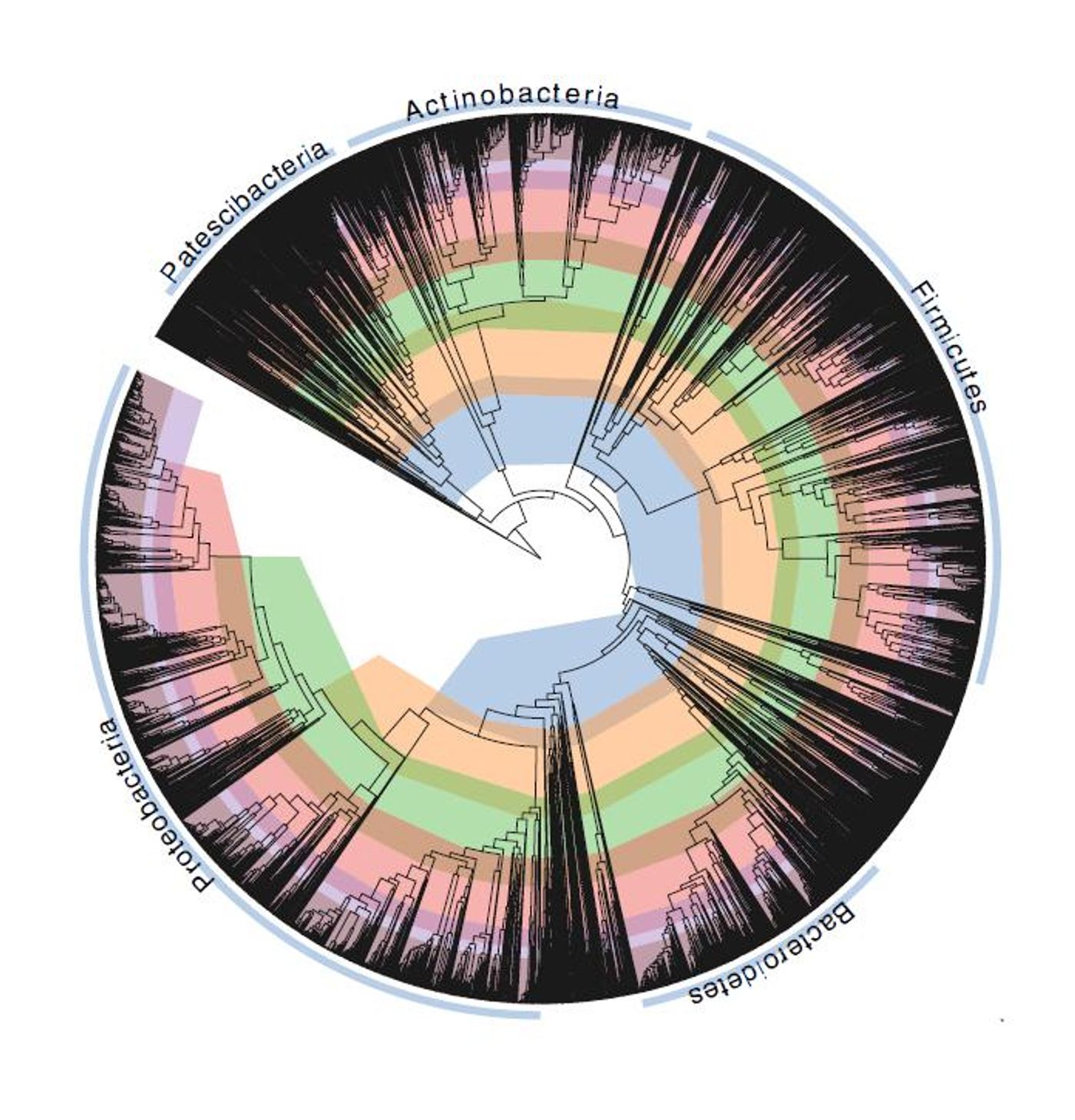
The classification of organisms into groups, taxonomy, has taken a step forward thanks to work by researchers at the University of Queensland. Led by Professor Philip Hugenholtz, the team approached classification through genetics and reconstructed an evolutionary tree with metagenomics, in which bacterial genomes were sequenced using samples taken directly from the environment. Reported in Nature Biotechnology, this approach aims to generate a more accurate, complete spectrum of data representing the true structure of bacteria’s tree of life.
This taxonomic research, said Hugenholtz, helps understand how living things are connected. "Taxonomy helps us classify living things by arranging them in a hierarchy from closely to distantly related organisms according to ranks, such as species, genus, family, order, class, phylum, and domain," he explained. "It's a system that helps us understand how organisms are related to each other, just like we do for time - using seconds, minutes, hours, and so on - or for geographic locations, using a street number, street, suburb, state, and country."
Professor Hugenholtz, of UQ's School of Chemistry and Molecular Biosciences and the Australian Centre for Ecogenomics (ACE), said scientists tend to agree that evolutionary relationships are an obvious way to classify organisms, but there are many mistakes in bacterial taxonomy because of historical problem
"This is mainly because microbial species have very few distinctive physical features, meaning that there are thousands of historically misclassified species," he added. "It's also compounded by the fact that we can't yet grow the great majority of microorganisms in the laboratory, so have been unaware of them until quite recently."
One team member, lead software developer Dr. Donovan Parks, credits exciting recent advances in genetic sequencing technology with helping to reframe the bacterial kingdom. "It's developed to a remarkable degree, and we can now get the entire genetic blueprints of hundreds of thousands of bacteria, including bacteria that have not yet been grown in the lab," he said.
The genomic data enabled the investigators to create an evolutionary tree of bacteria that was based on 120 genes that are commonly shared, or conserved, among bacterial species.
"This tree helped us create a standardized model, where we fixed all of the misclassifications and made the evolutionary timelines between bacterial groups consistent," Dr. Parks said. "For example, the genus Clostridium has been a dumping ground for rod-shaped bacteria that produce spores inside their cells, so we reclassified this group into 121 separate genus groups across 29 different families. We've given bacterial classification a complete makeover, and we're delighted that the scientific community is just as excited about this as we are."
In the video, Hugenholtz discusses his research into what has been nicknamed microbial dark matter - unknown or unnamed bacterial species. His work builds on basic research that established microbiology as an important field; when scientists learned that DNA is shared as a basic unit of life among all organisms on earth, they knew that microbes were an essential part of the tree of life, and worth studying.
Sources: AAAS/Eurekalert! Via University of Queensland, Nature Biotechnology