A New Kind of Regulatory Element in the Genome
The common, two-stranded helical structure of DNA was discovered over one hundred years ago. Many researchers would go on to find that DNA molecules could form other types of secondary structures, like the four-stranded G-quadruplex (G4). In the late 80s, scientists began to find evidence that G4s could form in cells although their exact purpose was not well understood. In recent years, scientists have found that many molecules can interact with G4 DNA, including transcription factors that help control gene expression.
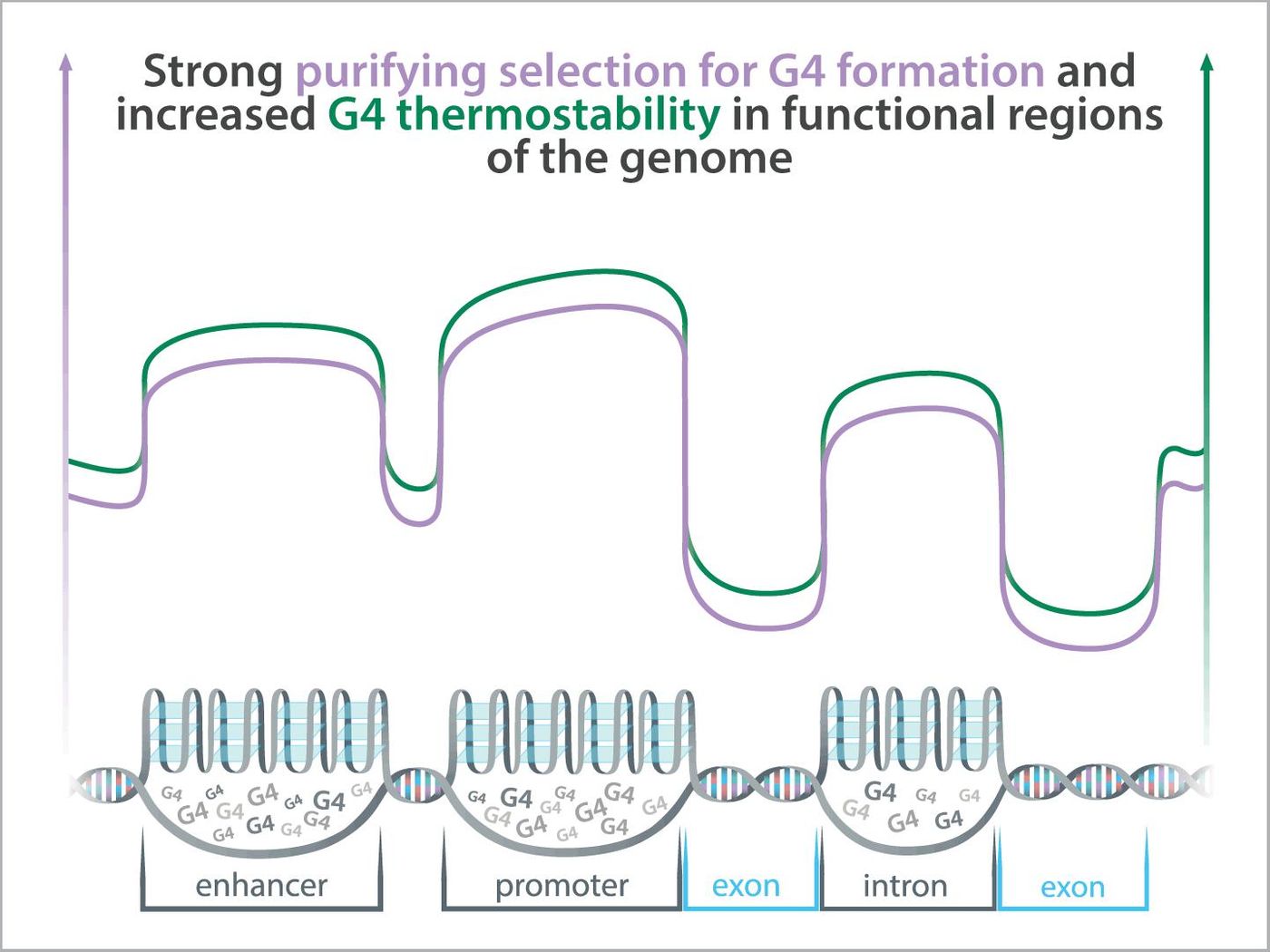
Scientists have proposed that regions of DNA where G4s tend to form are kept in the genome through natural selection, and that G4s play crucial roles in regulating gene expression. A new study has suggested that G4 elements that lie within regulatory regions of the genome that are functional but do not code for protein are more stable and can be found more frequently than G4 elements that sit within protein-coding genes. The work, which was reported in Genome Research, has suggested that regions that form G4 structures are another functional part of the genome, like non-coding RNA, for example.
"There have been only a handful of studies that provided experimental evidence for individual G4 elements playing functional roles," noted first study author Wilfried Guiblet, Ph.D. "Our study is the first to look at G4s across the genome to see if they show the characteristics of functional elements as a general rule."
It's been estimated that one percent of the genome is capable of generating a G4 structure (less than 2 percent of the genome codes for protein). G4 regions carry a lot of guanine, the 'G' nucleotide base. A variety of studies have found associations between G4 DNA and normal cellular processes and well as diseases including cancer.
"The three-dimensional structure of G4s can form transiently and how stable their structure is depends on their underlying DNA sequence and other factors," said Guilbet. "We found that usually, G4s located within functional regions of the genome tend to be more stable. In other words, it's more likely that the DNA is folded into a G4 at any given time and thus, more likely that the G4 is there for a functional reason."
Important parts of the genome have to be maintained; genes that are involved in crucial processes don't change much over time, and often are similar from one species to another. These regions are said to be kept up by purifying selection, which acts to eliminate mutations that arise in these genetic sequences. But other parts of the genome are more amenable to change; mutations in these sequences may not have any negative biological impact, and are thought to evolve neutrally. G4 elements may be in either of those regions.
Recent work by this team has suggested that G4s have high rates of mutation, and G4s are maintained by purifying selection even when they sit outside of important, protein-coding regions. The researchers suggested that this indicates they are necessary functional elements.
"We can look at the patterns of change in a DNA sequence among human individuals and between humans and our close primate relatives as a test of natural selection and then use selection as an indicator of function. Our tests show that G4s located within functional regions of the genome appear to be under purifying selections, which is further evidence that G4s should be considered as functional elements," explained study co-leader Yi-Fei Huang, an assistant professor of biology at Penn State. "The only exception from this pattern were protein-coding regions of genes, where G4s are relatively uncommon, rather unstable, and do not evolve under purifying selection. G4s in protein-coding regions of genes might be nonfunctional and costly to maintain."
"We think that we are seeing evidence for a paradigm shift for how scientists define function in the genome," said study co-leader Kateryna Makova, the Verne M. Willaman Chair of Life Sciences at Penn State. "First, geneticists focused almost exclusively on protein-coding genes, then we became aware of many functional non-coding elements, and now we have G4s and possibly other non-B DNA elements. Three-dimensional structure may be just as important for defining function as the underlying DNA sequence."
Sources: AAAS/Eurekalert! via Penn State, Genome Research