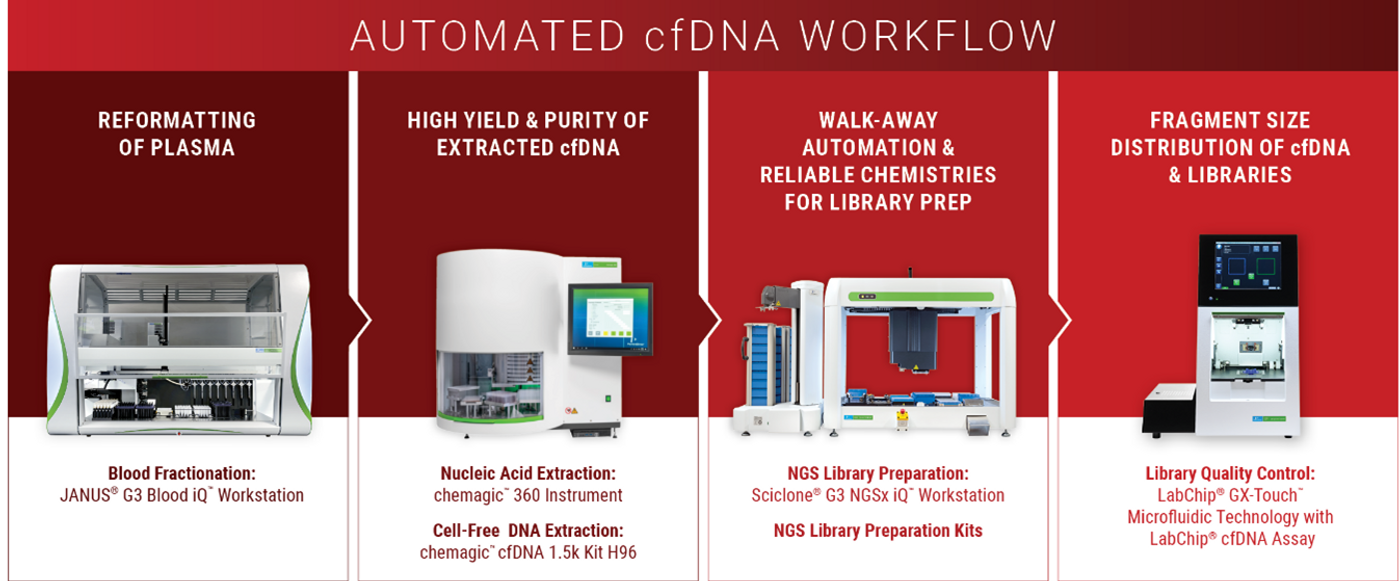
Tips on Establishing a Reliable Cell Free DNA Workflow

Sample Collection
It is key during sample collection to minimize genomic DNA contamination that can arise from white blood cell (WBC) lysis. Hence, plasma is preferred to serum as WBC lysis occurs during clotting.
Recommendations:
- Minimize cell lysis during blood drawing e.g., use appropriate needle size, avoid prolonged tourniquet
- Isolate plasma carefully, avoiding contact with buffy coat layer (automatable with PerkinElmer’s JANUS® G3 Blood IQ™ Workstation)
- Isolate plasma within 6 h of blood collection when using EDTA blood tubes, or longer for specialized blood collection tubes containing stabilizers (see manufacturer’s instructions)
- Perform a double centrifugation of plasma to minimize carryover of WBCs
Extraction of cfDNA from Plasma
Studies comparing commercially available kits have reported varying yields and quality, but results may be influenced by variabilities in sample source, handling steps and quantification procedures across labs.
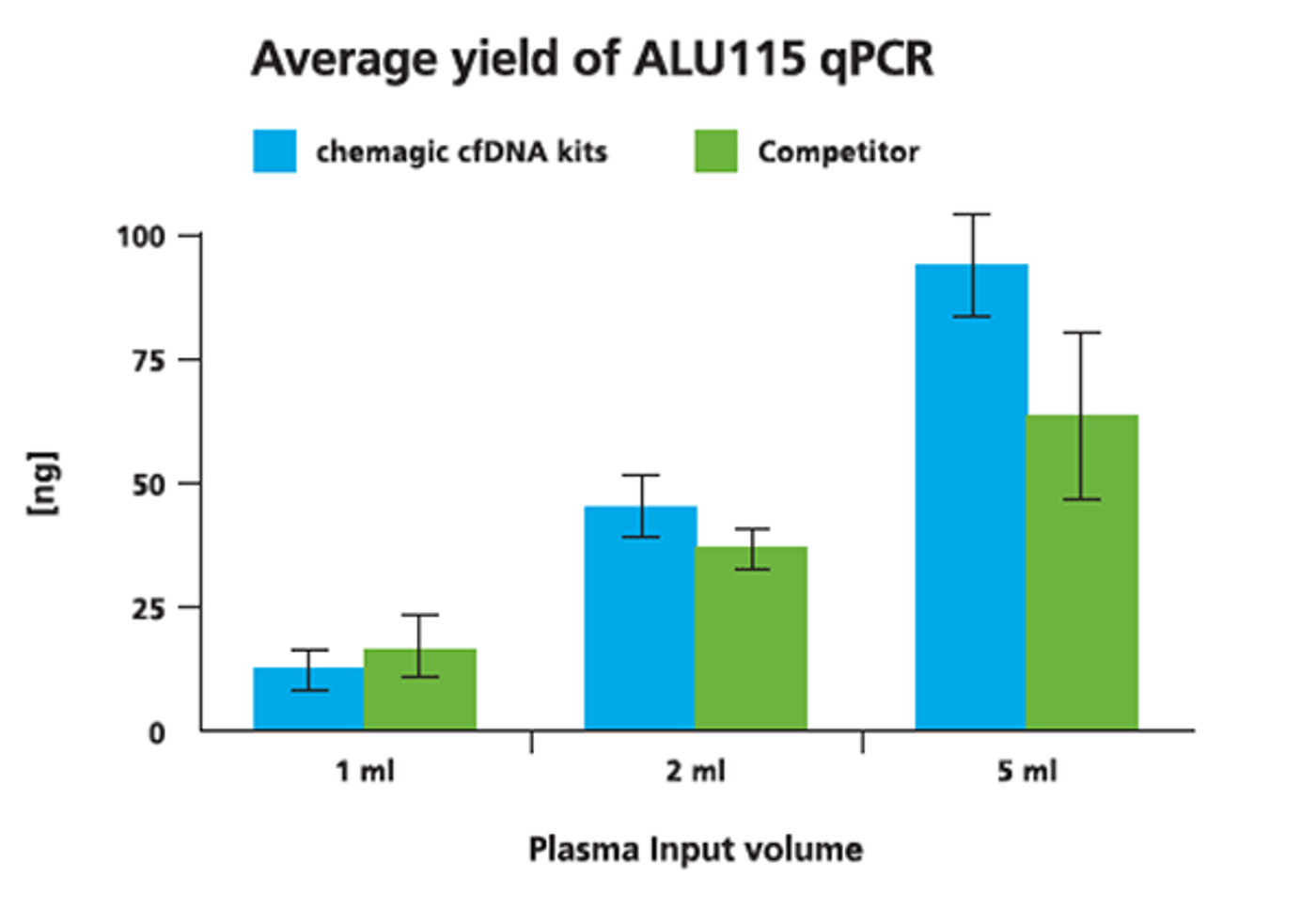
We tested the chemagic™ cfDNA extraction kits, based on patented chemagic™ M-PVA Magnetic Beads, on the chemagic™ 360 instrument and obtained comparable and consistent yields to a competitor’s manual silica column and vacuum-based method.
As different extraction kits have reportedly variable extraction efficiencies based on DNA fragment length, we also analyzed fragmentation of obtained cfDNA by PCR using the KAPA™ Human Genomic DNA Quantification and QC Kit from Roche. Fragmentation/degradation scores with the chemagic extraction kits were in the range of (0.16 - 0.37) which was comparable to the results gained with the silica membrane-based competitor kit on the same samples (degradation scores 0.15 - 0.4).
cfDNA was isolated from 1 ml, 2 ml and 5 ml from two donors both with the chemagic kits on the chemagic 360 instrument and manually with competitor. For cfDNA analysis, a short fragment (115 bp) from a consensus sequence with abundant genomic ALU repeats was amplified. Exemplary data from donor 2 shows that the yield of cfDNA is scalable to sample input.
Aside from increasing consistency of cfDNA extractions, automation with chemagic™ technology can:
- Increase sample throughput
- Reduce hands-on steps and speed up workflow (< 2 h for up to 96 samples)
- Enable a wider sample input range (0.5 to 18 ml)
- Maintain sample integrity with sample tracking options
- Reduce running cost per sample
“The PerkinElmer cfDNA extraction kit on the chemagic 360 platform has greatly reduced the hands-on time needed for sample preparation. It has also given us improved cfDNA yields compared to the current, manual approach used in our laboratory and enabled significantly higher throughput. The ease-of-use of the system is high and has made it easy for us to rapidly train our staff and implement the approach in our day-to-day practice.”
Veli-Mikko Puupponen, CEO, BiopSense Oy/Ltd, Finland
Quantification and Quality Control of Extracted cfDNA
Before cfDNA is subject to downstream sequencing, quality control checks are often performed. As cfDNA yields from normal samples typically range between 1 - 30 ng/mL of plasma, this may fall below detection sensitivity of spectrophotometric methods. Hence, quantification by PCR (qPCR or ddPCR) is recommended.
Recommendations for cfDNA yield verification
- qPCR using an ALU115 primer set where a short fragment (115 bp) from a consensus sequence with abundant genomic ALU repeats is amplified
- other qPCR/ddPCR-based assays targeting “housekeeping” genes found in cfDNA which have been experimentally validated can be utilized
- the LabChip® cfDNA Assay is optimized to provide fast, automated qualitative and quantitative analysis of cell-free DNA (cfDNA)
For more detailed tips and references, read this Blogpost
For research use only. Not for use in diagnostic procedures.