Bacteria Seen Stealing Resistance Genes During Combat
Scientists at the University of California San Diego Center for Microbiome Innovation have figured out one way that an important bacterium gains resistance to antibiotics. They have used their work to create a model that can predict the conditions that help the resistance to spread as well. The research establishes a paradigm for investigating, measuring and potentially, fighting the rise of antibiotic-resistant superbugs. The data has been reported in the open-access journal eLife.
Researchers are trying to learn more about pathogens that are able to evade drug treatment, and these dangerous microbes are becoming a significant threat to public health. The Acinetobacter species of microorganisms are among the most severe resistant pathogens; the bacteria love to grow in hospitals and easily pick up genes that provide them with drug resistance through a mechanism called horizontal gene transfer.
"Most of the threat posed by Acinetobacter stems from its ability to acquire drug resistance via horizontal gene transfer (HGT)," said the senior author of the report, Jeff Hasty, professor of biology and bioengineering at UC San Diego.
Bacteria can exchange genetic material using HGT, and the Acinetobacter species can do so very well. That makes them excellent at gaining resistance to drug treatments. HGT had not been seen directly in Acinetobacter species, but serendipitously, the investigators did so.
Robert Cooper started his postdoctoral work in the Hasty lab trying to use algae and bacteria to produce biofuels.
"I was looking for a side project and remembered reading about 'predator' Acinetobacter bacteria that engage in hand-to-hand combat to kill their prey," explained Cooper. "They do so by extruding something analogous to a poison-tipped dagger, and stabbing neighboring cells."
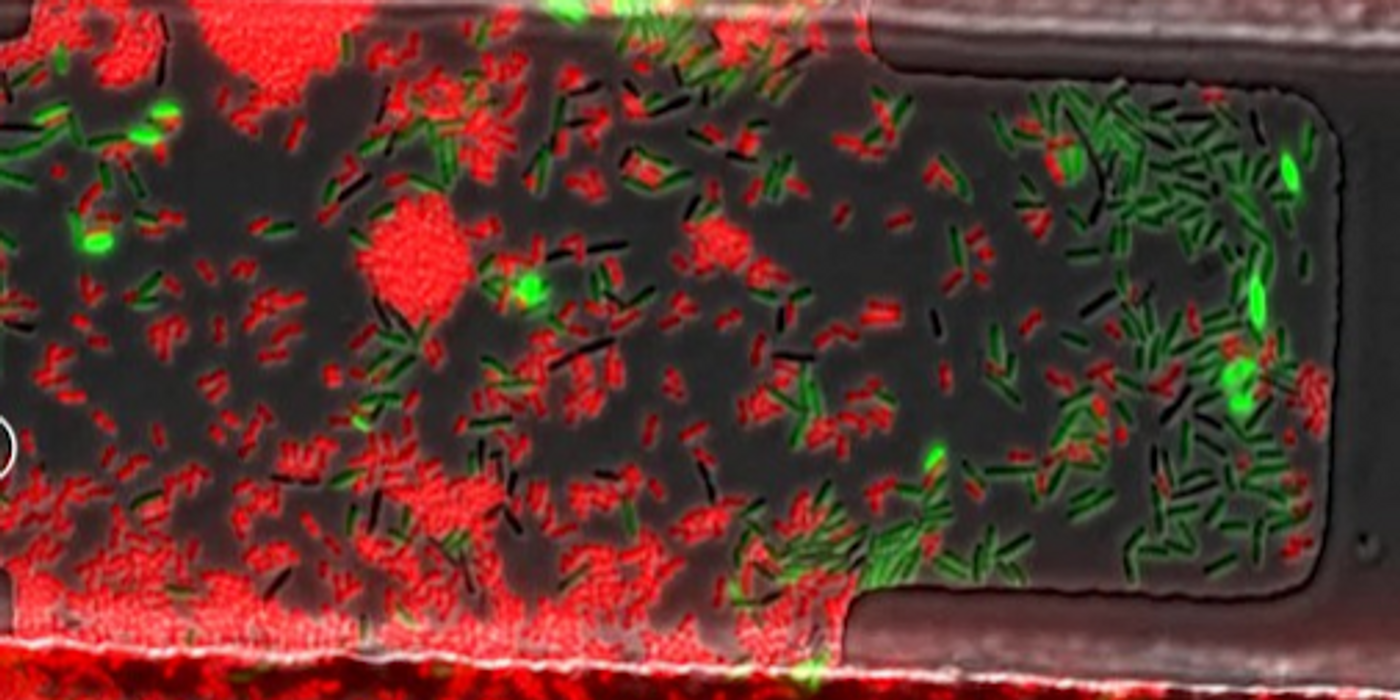
Cooper was intrigued, so he acquired some predatory bacteria, then grew them adjacent to E. coli cells that acted as a prey. Each was given a fluorescent protein; the predators got red, and the prey got green so they could easily be distinguished from one another in movies of their activity. Indeed, the bacterial predator killed its E. coli prey as expected, but then the predators began to glow green!
The video shows cells growing inside a tiny chamber. Each oval is one bacterium.
Cooper's research then took a new turn. After attaching a green fluorescent protein to a gene that confers antibiotic resistance, he could see that when Acinetobacter killed its prey, it took up the green gene, and also acquired the antibiotic resistance gene, which it incorporated into its genome. The video illustrates the process. A colony of drug-resistant Acinetobacter was born.
Next, a model was created by Cooper to predict how many prey cells would be destroyed, and how many predator cells would then gain the antibiotic resistance gene under various conditions.
"The model predicts the efficiency of uptake and incorporation of the antibiotic resistance gene," said Cooper. "We hope that it will be useful for determining the conditions under which antibiotic resistance might spread, which could help us focus on the most important areas for prevention."
Sources: Phys.org via UCSD, eLife