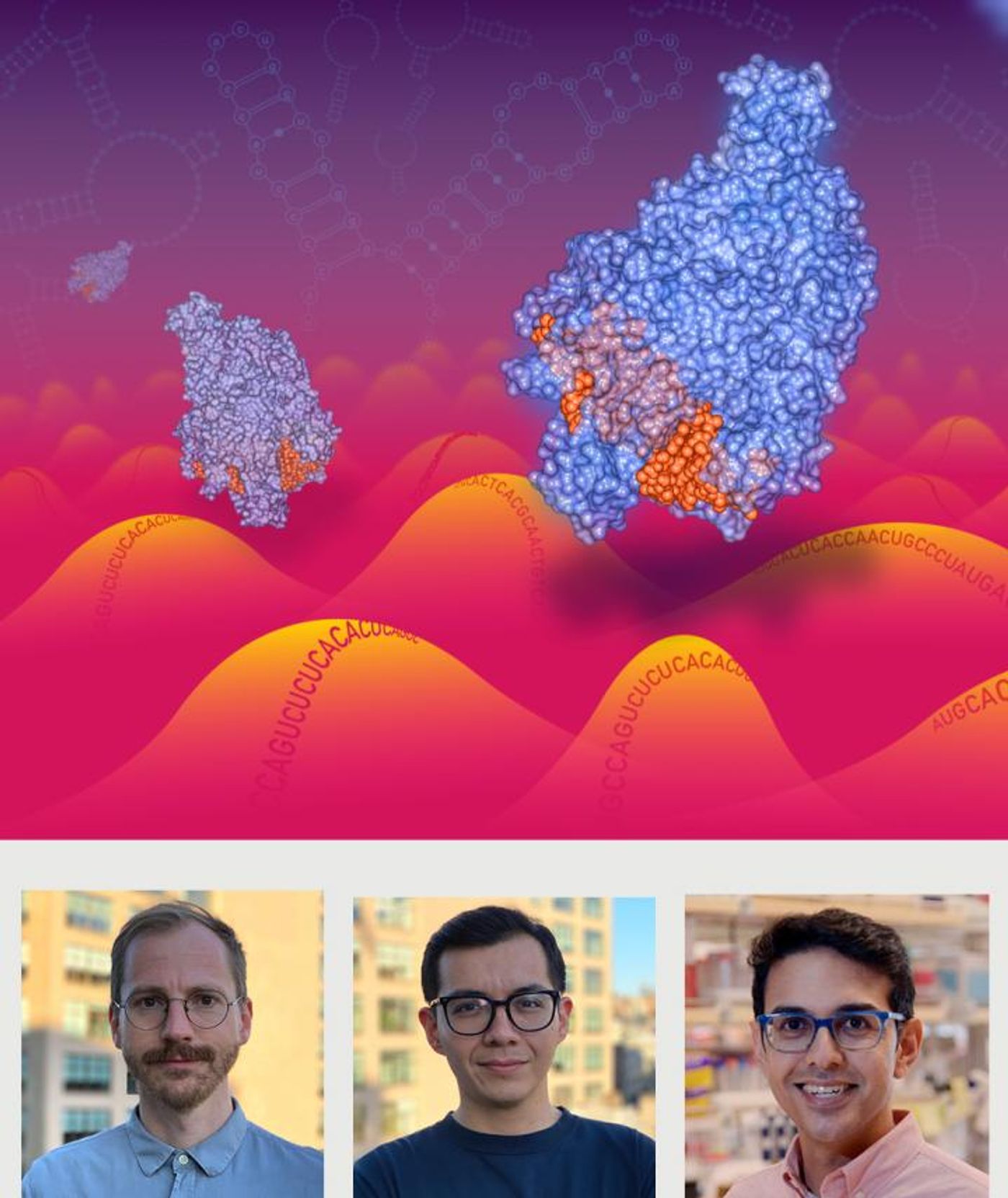
CRISPR is well-known as a gene-editing system, but it can be used for other purposes as well. Screens that employ CRISPR have revealed genes that are involved in disease, like sickle cell anemia or the metastasis of lung cancer, but these assays are aimed at DNA. For treating other illnesses, it may be necessary to target a different molecule, called RNA. New research has used a type of CRISPR enzyme called Cas13 to target RNA instead of DNA.
RNA is typically known as a kind of intermediate molecule that is used to carry the genetic instructions for making a protein from the genome to the cell's protein-making machinery. Other kinds of non-coding RNA can play regulatory roles and help control when genes are expressed.
In this research, the Cas13 enzyme was utilized in a high-throughput screen. These can now be used to learn more about RNA regulation and how non-coding RNAs are working. Thousands of RNA transcripts were targeted by the investigators in different ways, and the scientists found which targets were most effective. The data is now open to other researchers, who can use the pre-designed guide RNAs to target any protein-coding human gene.
"We anticipate that RNA-targeting Cas13 enzymes will have a large impact on molecular biology and medical applications, yet little is known about guide RNA design for high targeting efficacy," said the senior author of the study Dr. Neville Sanjana, Ph.D., at the New York Genome Center, among other appointments. "We set about to change that through an in-depth and systematic study to develop key principles and predictive modeling for most effective guide design."
The CRISPR (clustered regularly interspaced short palindromic repeats) / Cas (CRISPR-associated proteins) system is part of a bacterial defense system. Scientists have used the process to target specific places in the genome with a special RNA molecule, along with an enzyme like Cas9 that then makes a cut. But Cas13 enzymes are guided to RNA molecules and can make edits there instead, thereby altering gene expression but not changing the genome of a cell permanently.
This study analyzed how about 24,000 guides targeting RNA were working. "We tiled guide RNAs across many different transcripts, including several human genes where we could easily measure transcript knock-down via antibody staining and flow cytometry," said postdoctoral scientist Hans-Hermann Wessels. "Along the way, we uncovered some interesting biological insights that may expand the application of RNA-targeting Cas13 enzymes."
They identified a critical part of the target RNA that is especially sensitive to mismatches between target and guide. This knowledge will help researchers to reduce unintended edits, so-called off-target activity. This area may also help scientists design biosensors that can detect small differences between RNAs that are very similar.
"We are particularly excited to use the optimized Cas13 screening system to target noncoding RNAs," said study co-first-author Méndez-Mancilla. "This greatly expands the CRISPR toolbox for forward genetic and transcriptomic screens."
This work has also helped identify an RNA that could be used in the detection and potentially, treatment of COVID-19 in the future.
Sources: AAAS/Eurekalert! via New York Genome Center, Nature Biotechnology